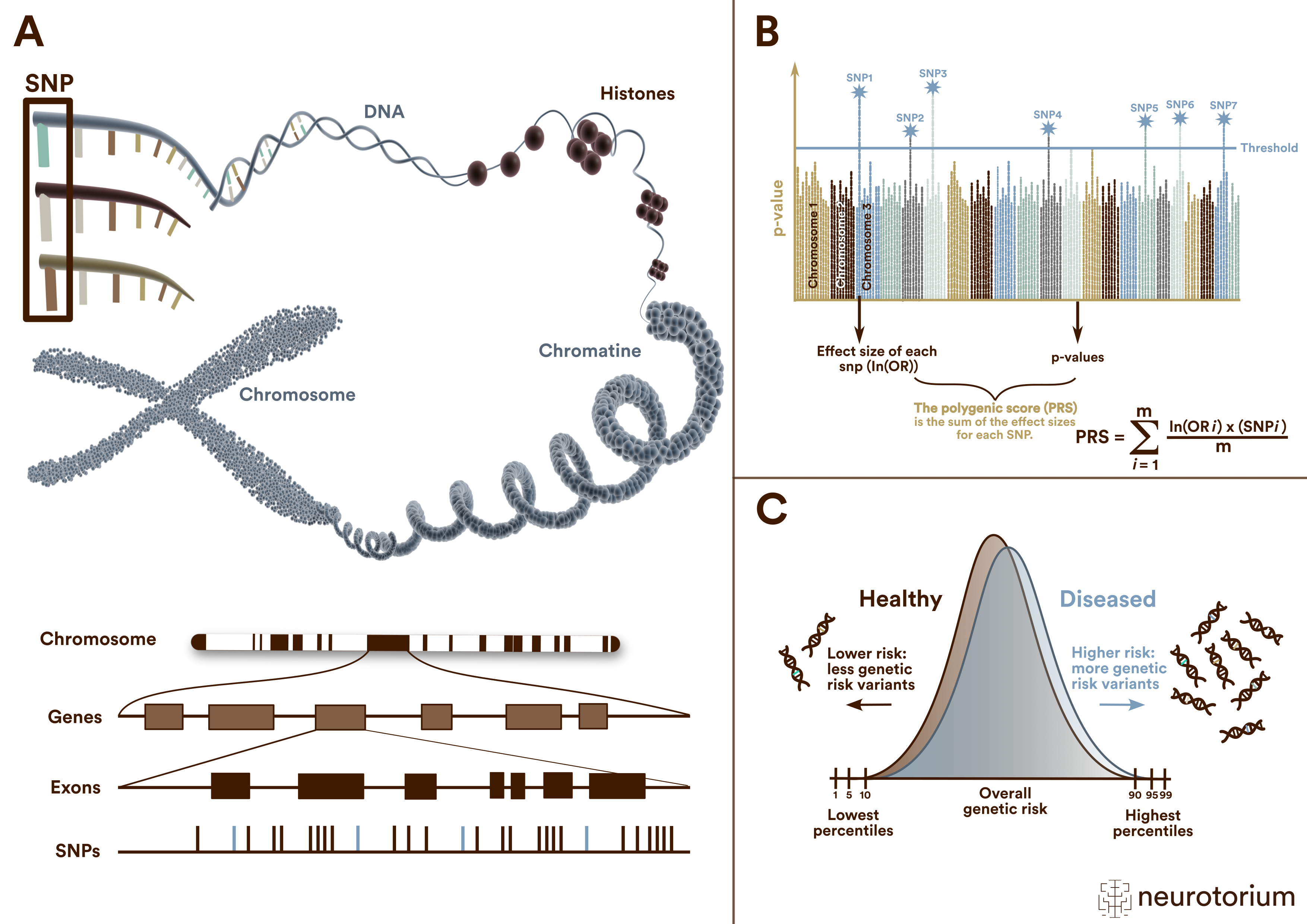
(A) A single nucleotide polymorphism (SNP) is a frequent (>1%) genomic variant at a single base position in the DNA. Some SNPs in the coding region of a gene (exon) change the amino acid sequence of a protein, and others in the coding region do not affect the protein sequence.
(B) Polygenic risk scores (PRS) are typically calculated by summing the weighted allele counts of independent (i.e., uncorrelated) SNPs found to be associated with a trait or disorder in a genome-wide association study (GWAS). For quantitative traits, the weights are the linear regression effect sizes, i.e., the β coefficients. The weights for case/control phenotypes are the natural logarithm of the odds ratio, ln(OR).
(C ) Polygenic risk scores (PRS) follow a normal distribution on a population level. The distributions of cases and controls clearly overlap. Thus, meaningful risk predictions can be only expected for extreme quantiles at the top and bottom of the PRS distributions.